Research
Fluorescence Single-Molecule Bio-Imaging Project
Naoki Watanabe
Since 2002 (revised and supplemented in 2011)
- I. Background
- II. Which way to go? : actin polymerization-depolymerization pathways
- III. Retrograde actin flow
- IV. Bridging the gap between cellular signaling and cytoskeletal reorganization
- V. Formin homology proteins: the processive actin polymerizers (new)
- VI. Dynamic equilibrium of G- and F-actin as a trigger of actin nucleation (new)
- VII. Visualizing the actions of small compounds (new)
Reference: (Recent review) Watanabe N. Proc. Jpn. Acad. B. Phys. Biol. Sci. 86: 62-83. (2010) Link 1, Link 2
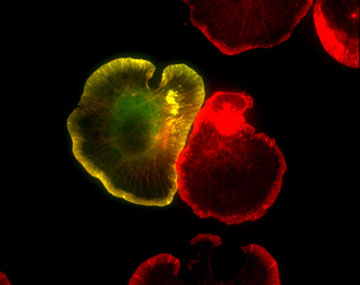
Figure 1: GFP-actin (green) and Texas red phalloidin staining (red) in spreading XTC fibroblasts (fixed). F-actin-rich pseudopods called lamellipodia are well developed at the cell edge of spreading fibroblasts.
I. Background
The human genome is finally decoded, and life science is advancing as vigorously as ever. Signal transduction research is not an exception. A number of molecular mechanisms have been found, and their genetic and biochemical interplay are continuously being revealed. However, much remains uncertain about what is really going on within living cells. Our knowledge is often diagramed in a scheme far simpler than in the real in-vivo situation. This reflects our lack of ability for observing on-going processes.
To overcome such deficiency, I developed the fluorescence single-molecule speckle method which allowed visualization of actin polymerization and depolymerization at the single-molecule level (Watanabe N. and Mitchison T. J., Science 295: 1083, 2002). With this method, we were able to measure the actin polymerization-depolymerization kinetics at the leading edge of living cells with high precision. The data revealed that more actin polymerizes in the body of lamellipodia than at the cell edge, and that actin filaments start depolymerization within seconds after polymerization (See Figure 2 and Movie 1-2). Then, what does this fast actin turnover in cells imply?
My laboratory is working on the following issues to understand the basis of cell migration, and trying to establish the next level of science capable of proving molecular processes directly in live cells.
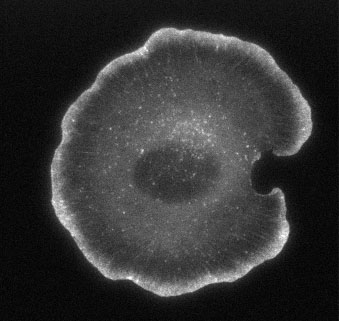
Figure 2. EGFP-actin in a live, spreading XTC cell, and its timelapse movie (Movie1).
Movie 1. EGFP-actin expressed at a high level (conventional GFP-actin movie). Total time, 10 minutes. The centripetal flow of the bulk actin network is observable.
Movie 2. Single-molecule actin speckles in a live XTC cell.The expression level of EGFP-actin is hundreds times less than in the cell in Fig. 2. This low density labeling enables visualization of individual molecules bound to cellular structures. Individual fluorescent actin copolymerized with cellular filaments is clearly seen. Time is in minute:second.
II. Which way to go? : actin polymerization-depolymerization pathways
Actin polymerization generates force that pushes the cell edge forward. To ensure fast polymerization, actin filaments do not stay long; depolymerization must be fast too. These notions are supported by the rapid loss of leading edge dynamics (within 1 minute) upon treatment with cytochalasin, an actin elongation inhibitor, or jasplakinolide, an actin depolymerization inhibitor.
But even in test tubes with purified components, actin polymerization and depolymerization pathways are not readily determined. Especially, under the conditions within lamellipodia (a ~200 nm thick veil-like structure at the cell edge) where actin exists at a 1000 micromolar concentration with densely packed regulatory proteins, it is of particular interest to see what such extreme conditions deliver.
By extending our single-molecule speckle approach not only to actin but also to actin regulatory molecules, we are now trying to clarify how actin polymerization and depolymerization proceed in live cells, and to raise the veil over the signals by which the cell protrusion machinery regulates actin.
III. Retrograde actin flow
The actin network at the cell periphery continuously moves toward the central region (retrograde actin flow). Although the centripetal movement of cell peripheral structures was recognized from observations using bright field microscopy as long ago as the early 1970s, it is still largely unknown how and why this massive actin flow is generated. Actin is believed to push the leading edge forward by its polymerization. However it also moves backward toward the center.
Presumably the actin flow is, in part, driven by certain members of the myosin family. Another important theory is the ‘clutch model’ in which the retardation of the actin flow by the strengthened connection of the actin network to the extracellular matrix has been postulated to lead to cell edge protrusion.
Now our single-molecule speckle method allows simultaneous measurements of actin polymerization distribution and the actin flow rate change accompanied by cell protrusion. We hope to identify the mechanisms closely connected with the actin flow and to elucidate the basic nature of the cell protrusion machinery.
IV. Bridging the gap between cellular signaling and cytoskeletal reorganization
We are also trying to apply the single-molecule speckle method to a signaling molecule, namely mDia1, a Rho effector. Since the intramolecular interaction of mDia1 is disrupted upon the binding of Rho-GTP (Watanabe N. et al., Nat. Cell Biol. 1999), and mDia1 presumably binds actin filaments afterwards as is the case of other Formin-related proteins, we expect to capture on-going Rho-mDia1 signaling.
Indeed we are capturing images of mDia1 dynamically associating with the cellular actin cytoskeleton and acting on it. From these new imaging experiments, we hope to both map the spatial and temporal nature of Rho-mDia1 signaling in live cells and visualize the actin reorganizing process conferred by this signaling.
V. Formin homology proteins: the processive actin polymerizers (new)
Formin homology proteins (Formins) play essential roles in cytokinesis and cell polarity formation throughout eukaryotes. Our single-molecule approach has revealed the processive actin elongation property of Formins (Higashida C. et al., Science 2004). By continuously associating the fast growing end of an actin filament, Formins facilitate the long-range actin elongation, which is presumably prerequisite for the formation of actin stress fibers and yeast actin cables. Moreover, in cooperation with profilin, Formins accelerate actin elongation several fold beyond the theoretical diffusion limit of monomer associating to the filament end. In the cell, an mDia1-bound barbed end incorporates 720 actin subunits per second (See Movie 3).
Recently by observing single-molecule fluorescence polarization, we demonstrated rotation of mDia1 along the helical twist of an assembling actin filament (Mizuno H. et al., Science 2011). Formins might destine cellular actins for elongation or disassembly by imposing torsional stress around the filament axis.
Movie 3. Single-molecule imaging of actin polymerization-driven molecular movement of the mDia1 FH1-FH2 domain mutant, ΔN3. mDia1 directionally moves at ∼2 μm/sec by processively polymerizing an actin filament until being trapped by the actin network or the cell leading edge. Time is in second. Scale bar, 2 μm.
VI. Dynamic equilibrium of G- and F-actin controls actin nucleation by Formins (new)
In search of compounds that activate mDia1, we unexpectedly found paradoxical activation of mDia1 by an actin monomer inhibitor, latunculin B. Pharmacological kinetic simulation revealed that latrunculin increases polymerization-competent actin monomers in the complex actin turnover circuit. This ‘latrunculin paradox’ is an unprecedented case of an increase in the drug-free target induced by the drug. Notably, mDia1 frequently nucleates actin filaments around the high actin depolymerizing foci in the cell (Higashida, C. et al., J. Cell Sci. 2008). Rapid perturbation of the system by small compounds and its quantitative modeling thus shed light on the role of the dynamic equilibrium in complex biological systems.
VII. Visualizing the actions of small compounds (new)
We also apply our single-molecule approach to elucidation of the mechanisms of action of target-based drugs. Recent findings including our own (Fujita A. et al., Mol. Pharmacol. 2009) suggest that inhibitor binding to the ATP pocket may lead to conformational change of the kinases. This notion will give a clue to develop a new strategy for drug discovery.